Now how do I get it do this on a full study instead of a run where the sra files are in a crap load of folders? I am on a 32 bit windows system running XP I know, really dated. National Center for Biotechnology Information , U. You should enable this flag as it will trim off those sequences for you. Use with "X" to dump a range. 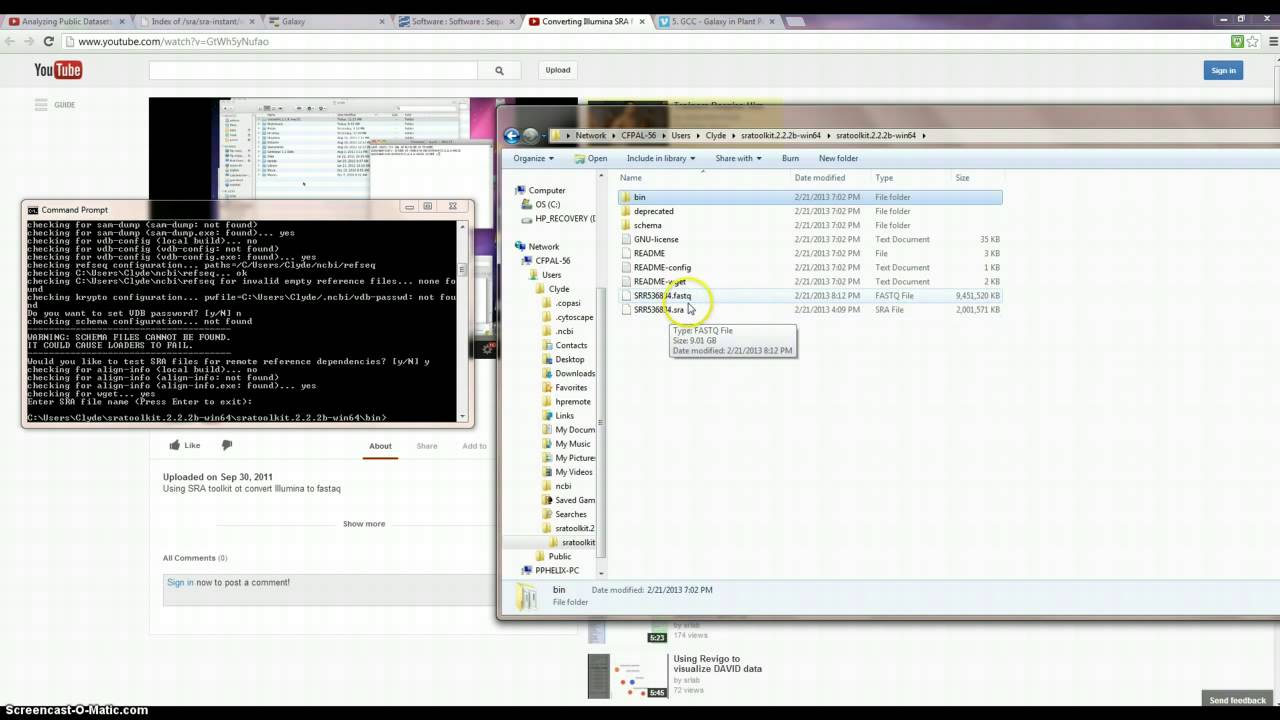
| Uploader: | Jusar |
| Date Added: | 18 October 2012 |
| File Size: | 7.63 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 20490 |
| Price: | Free* [*Free Regsitration Required] |
Generally this will put you in your "home directory" e.
This will ensure that your output has A, T, G, and C instead of being put into color space. Support Center Support Center. Originally Posted by colindaven. Data submitted as aligned bam are output as aligned sam, while other formats are output as unaligned sam.
If you split your sequences into one using —split-spot or two using —split-files files, by default the sequences get the same ID. That makes no sense and will break dunp downstream processes because you have duplicate sequence IDs in the same file.
Converts data to sff format. Please send all Toolkit questions to: You are bound to run into some problem or the other using bit windows. By default the output is written to one or more files. Last edited by GenoMax; fstq Not sure if this would work.
fastq-dump for dummies - SEQanswers
This is optional and up to you. Send a private message to simonandrews.
Downloading from there is pretty simple and fast, see my tutorial on that: A lot of downstream software requires fasta sequences only, and this is a good way to get those sequences. Originally Posted by colindaven fastq-dump mySRA. You can also use the toolkit to convert from the formats listed below into the SRA format not required for submission, but will allow you to use the SRA Toolkit to archive or analyze your data:.
What is the purpose of the SRA toolkit? Unfortunately, it is not very well explained. What's your method to check the completeness of the fastq file after the download by fastq-dump from SRA database?
NCBI SRA file format - Metagenomics
If you do have access to a bit machine and NTFS formatted disks that can handle large single files duump may want to switch. Some of the reads in SRA are paired-end reads where they sequenced e. Extract the toolkit software folder and place it into a suitable location. Therefore, a header that starts out:. The default parameters for fastq-dump are also ridiculous and certainly not what you want to use.

National Center for Biotechnology InformationU. Note that only data submitted as sff can be converted back to this format. Send a private message to UpsetNotMad Scientist. Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
fastq-dump
Apparently the error you mentioned may be related to absence of service pack 3 for XP. Powered by Biostar version 2.
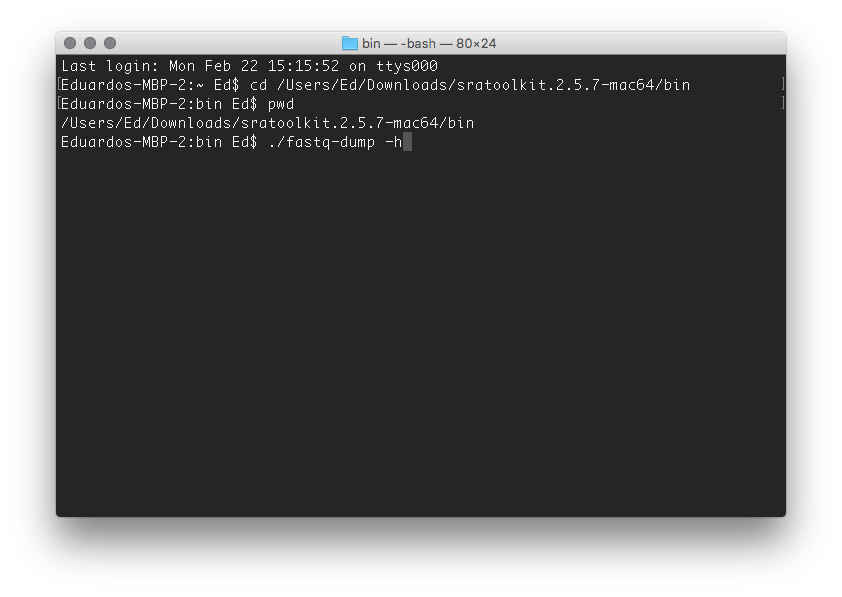
Still, you can get most data directly from the European Nucleotide Archive in fastq format. If you are working with SRA files you will need, at some point, to use fastq-dump.

Комментарии
Отправить комментарий